Study of quinones biosynthesis (Fabien Pierrel, Ludovic Pelosi, Ivan Junier, Sophie Abby)
We combine biochemical, genetic and evolutionary approaches on model micro-organisms to advance our molecular understanding of the biosynthesis and function of respiratory chains, which are central to cellular bioenergetics. Indeed, respiratory metabolism, which results in the complete oxidation of nutrients to CO2 and the concomitant synthesis of ATP, provides most of the energy of eukaryotic cells and many bacteria. As such, dysfunction of respiratory metabolism has severe cellular consequences and leads to mitochondrial diseases in humans.
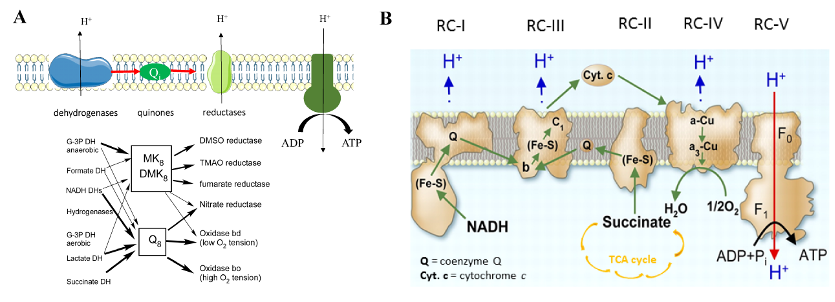
Bacterial and eukaryotic mitochondrial respiratory chains are composed of several modules, among which quinones constitute an essential link between primary dehydrogenases and terminal reductases (Figure 1).
 |
|
Figure 1: Bacterial (A) and mitochondrial (B) respiratory chains. Quinones (Q and (D)MK) transfer electrons (arrows) between respiratory enzymes, which generate a proton gradient used by ATP synthase to synthesis ATP. |
Eukaryotes possess only ubiquinone, also named coenzyme Q (Q), whereas most proteobacteria contain both Q and menaquinone. Q was discovered almost 60 years ago, yet several aspects of its biosynthesis remain obscure and notably the identity of proteins catalyzing several reactions. We are currently involved in the following research projects:
1) Identification and characterization of genes contributing to coenzyme Q metabolism:
 |
|
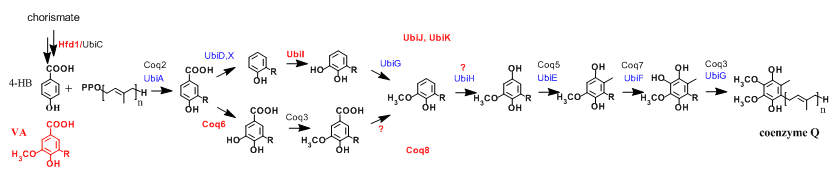
Figure 2: Coenzyme Q biosynthetic pathways in E. coli (protein names in blue) and S. cerevisiae (black), n=6-8. Significant contributions of the group are in red (Coq8, UbiJ and UbiK participate to Q biosynthesis without catalyzing chemical reactions). 4-HB, 4-hydroxybenzoic acid and VA, vanillic acid. |
|
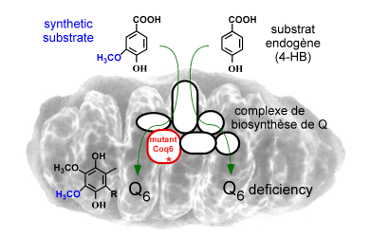
Over the past 10 years, we have contributed to identify new genes required for Q biosynthesis and to uncover their molecular function by studying mainly the yeast Saccharomyces cerevisiae and the gamma-proteobacteria Escherichia coli and Salmonella typhimurium (1-6) (Figure 2). In the latter organism, we found that Q is important for efficient bacterial proliferation inside macrophages pointing to the Q biosynthetic pathway as a potential antibacterial target (3). In S. cerevisiae, we discovered that synthetic analogues of 4-hydroxybenzoic acid, the precursor of Q, can enter the Q biosynthetic pathway and restore Q deficiencies linked to impairment of given hydroxylation reactions (7,8) (Figure 3). These results have opened new perspectives for the treatment of primary coenzyme Q deficiencies in humans (9,10). |
|
|
|
Figure 3: The biosynthesis of Q is restored by vanillic acid (a synthetic analog of the endogenous 4-HB substrate) in S. cerevisiae cells carrying mutant coq6 alleles. |
2) Regulation and supramolecular organization of Q biosynthesis in E. coli
This project is developed in collaboration with the groups of Frédéric Barras (Marseille) and Marc Fontecave (Paris) and is supported by the national research agency (project “(An)aeroUbi“, grant number ANR-15-CE11-0001).
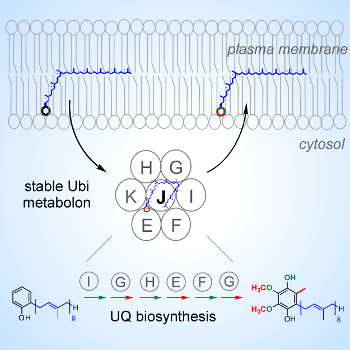
E. coli is a facultative anaerobic proteo-bacterium that can grow under anaerobic (absence of dioxygen) and aerobic conditions. Q is known to contribute to the aerobic metabolism of E. coli but is also produced in anaerobic conditions via a pathway that has been only partly elucidated.
|
|
||
|
Figure 4: The Ubi metabolon composed of seven Ubi proteins catalyzes the last six steps of the biosynthesis of coenzyme Q in E. coli (12). |
Overall, our characterization of the supramolecular organization and dynamics of Q biosynthesis in response to varying oxygen tensions will reveal important aspects of microaerobic and anaerobic metabolisms of facultative anaerobic proteobacteria.
3) Evolution of the regio-selectivity of Q hydroxylases in proteobacteria
This project is developed in collaboration with Ivan Junier and Sophie Abby from GEM team.
|
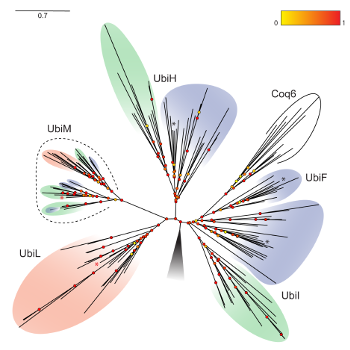
We recently revealed two new clades of Q hydroxylases (UbiM and UbiL) (Figure 5) and discovered that proteobacteria evolved a wide variety of solutions to catalyze the three hydroxylation reactions necessary for Q biosynthesis: some bacteria have a single hydroxylase with broad regio-selectivity (it can hydroxylate several positions of the aromatic ring), while others have three “specialist” hydroxylases with narrow regio-selectivity (they hydroxylate only a specific position) (13). We are now analyzing and clustering the Q hydroxylase sequences from all proteobacterial genomes available to reveal motifs linked to the regio-selectivity and thus obtain an understanding of the molecular evolution of these enzymes. |
 |
|
|
Figure 5: Molecular phylogeny of Q hydroxylases in alpha-, beta- and gammaproteobacteria (pink, green, and blue, respectively). |
Job and internship opportunities:
Come work with us in a fun and productive atmosphere! We are always willing to work with motivated students and postdoc interested in biochemistry, metabolism and evolution. For any project, applicants should contact Fabien or Ludovic by e-mail (fabien.pierrel [at] univ-grenoble-alpes.fr or ludovic.pelosi [at] univ-grenoble-alpes.fr).
Adaptation of lipid composition of bacterial membranes (Corinne Mercier)
Understanding how pathogenic bacteria such as Pseudomonas aeruginosa (the causative agent of lung infections linked to cystic fibrosis) and Escherichia coli (involved in gut and bladder infections) adapt to their environment is a key challenge for the development of new antibiotics. Membrane dynamics and homeostasis are essential mechanisms for bacterial survival and an understudied research area. We investigate how bacteria react to environmental changes by adapting the lipid composition of their membranes (lipidomic approaches). To identify novel therapeutics targets, we search for the mutations, which lead to these adaptations (genetics, cellular and biochemical approaches). This project, which addresses, for the 1st time, the question of how membrane lipids are modified in these versatile pathogens in response to environmental changes and degree of virulence, will provide data of both fundamental and clinical interest.
Selected publications: